
If you are interested in participating this Coordination Action please contact us!

In 2001, we initiated the Central Aspergillus Data Repository (CADRE). This project aims to support the international Aspergillus research community by gathering all genomic information regarding this significant genus into one resource. Using the Ensembl suite, CADRE has been publicly available since 2004. It currently facilitates the visualisation of annotation and comparative data (courtesy of Ensembl Genomes) for nine Aspergillus genomes. Currently, we are also involved in a European systems biology study of the specificity of response of the cell-mediated immune system to fungal microorganisms in order to investigate the genetic basis of susceptibility to fungal disease and elucidate molecular mechanisms of drug resistance in fungal pathogens. Our first test cases to arise from this project are nine different strains of Aspergillus fumigatus: CEA10, AF300, F17999, F18329, F18454, F20451, F21572, F21732 and F21857.
Aspergillus is a genus of moulds that are found world-wide. Over 300 species of it have been recognised, a small number of which cause illness in animals (including humans): A. fumigatus, A. flavus, A. terreus and A. niger. Most humans are naturally immune and do not develop any of the diseases caused by these moulds. However, when disease does occur, it takes several forms, ranging from an allergy-type illness to life-threatening systemic infection. The severity of the infection is determined by various factors, but one of the most important is the state of the immune system of the person. The most frequently isolated of these moulds from immuno-compromised patients is A. fumigatus.
Setting up a pipeline to process un-annotated genomes is fraught with difficulties. In theory, given a set of related data files, a typical pipeline ties in different programs to detect repeats, to predict genes and to combine the data to provide the best gene model (see flowchart). These predicted genes can then be used to detect homologues in public databases and be embellished further. In practice, however, each file provided for a project can describe data attributed to different assembly components (sometimes with no assembly description). In addition, each piece of software produces its own variation of a standard format that cannot be used by the next program in the chain. For example, for one of our test cases (Aspergillus fumigatus CEA10), assembled sequences are provided by chromosome but other data (e.g., RNA-seq, DIP and SNP) are provided by supercontig. There is no assembly file describing the relationships between the components. Initially, this in not a problem, but it does cause difficulties further down the line when trying to improve gene models, to visualise all data within a genome viewer for further analyses and to upload data into a related database such as CADRE.
We used GeneMark-ES and Augustus for ab inito gene prediction; both required the use of different in-house programs to convert the output before it could be passed onto Evidence Modeller (EVM). The output from EVM again required conversion before being used with BLAST to find the best match within an Aspergillus database. Development of an in-house program was required to merge resultant data into gff3 format for the next step in the process.
The Ensembl suite uses the gff3, agp and fasta file formats for uploading data into an ensembl database system such as CADRE. However, as pointed out earlier we do not always have consistent assembly information, therefore, we may need to re-create such files from a reference genome. A new database is initiated within CADRE with FASTA sequence files and assembly data (i.e., agp files), where appropriate. EBI software (GffDoc.pl), can then be used to upload predicted genes from gff3 files. All in all, this has become a rather bloated and time-consuming process. 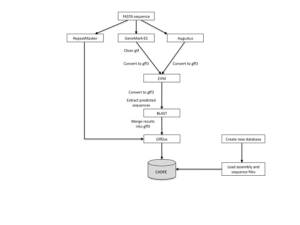 

To be able to use genome project data without worrying about the way in which it has been described. To enable the annotation pipeline to run without continual data reformatting.
As discussed in a previous section, we have written several in-house programs to try and sort out formatting issues. We would need to create a smoother process and have it less fragmented. We also need to develop gff, gtf and gff3 parsers that deal with more than just the two gene finding programs we have used. We would like to introduce GeneWise but this will probably have its own formatting issues so we would need to introduce flexibility.
The test case will have impact for the substantial worldwide fungal genomics community
LF: a typical pipeline to build (ideal test case!?) with probably a risk of failure in the long term run Invite people from Taverna or other such tools.